This pleiotropic transcription factor is involved in the regulation of cell survival, proliferation and differentiation in normal conditions, but also in pathological conditions as diverse as inflammation, neurodegenerative disorders, stroke, traumatic brain injury, radiation toxicity or cancer. We are studying the role of protein-protein interactions and post-translational modifications in its functions at a cellular level.
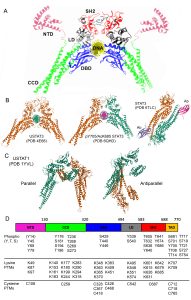
STAT3 domain organization and function
Signal transducer and activator of transcription (STATs) proteins constitute a family of SH2-containing transcription factors downstream the activation of various receptor-associated and non-receptor kinases by growth factors, cytokines and other signaling molecules. They are involved in a wide spectrum of physiological and pathological phenomena, including cell survival and proliferation, development and tissue morphogenesis, immunity, response to injury, neurodegeneration and cancer. Seven STAT genes have been described (i.e. STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B and STAT6), producing 7 distinct proteins plus a few alternative splicing variants and truncated forms. However, only deletion of STAT3 is lethal to mouse embryos.
STAT3 functions are largely controlled by PTMs and protein-protein interactions, and these are closely related to the amino acid sequence, domain organization and tertiary, 3-dimensional structure common to the STAT family of transcription factors (Fig. 1). The domains include, from the N-terminus to the C-terminus, an N-terminal domain (NTD), a Coiled-Coil domain (CCD), a DNA binding domain (DBD), a linker domain (LD), a Src homology region 2 (SH2) domain and a C-terminal transactivation domain (TAD)(Fig. 1). STATs are the only family of transcription factors with an SH2 domain, which allows them to bind phosphotyrosine residues in protein interactors, such as membrane receptors and other STAT molecules. NTD and SH2 domains are responsible for STAT dimerization and tetramerization and, together with the CCD, they mediate most interactions with other proteins. The DBD and LD bind to Interferon-Gamma Activated Sequences (GAS) in the DNA, enabling the transcription of specific target genes. Finally, the TAD is involved in fine regulation of STAT functions either by post-translational modifications (PTMs) or protein-protein interactions. The TAD contains the two tyrosine and serine residues that are currently considered the master regulators of STAT activity, which in the case of STAT3 are Y705 and S727.
The canonical STAT3 pathway
STAT3 signalling is frequently divided in canonical and non-canonical pathways. The canonical pathway involves an initial trigger by the binding of cytokines from the Interleukin-6 family to their membrane receptors and ends with the transcriptional activation of a specific set of genes. These cytokines include interleukins 6, 11, 27 and 31, Oncostatin M, leukemia inhibitory factor, ciliary neurotrophic factor, cardiotrophin-1, cardiotrophin-like cytokine and neuropoietin. However, it is also activated by other extracellular factors such as epidermal-growth factor (EGF), fibroblast-growth factor, transforming-growth factor b or retinoic acid, among others. Activated membrane receptors are tyrosine-phosphorylated in their cytosolic domains and recruit STAT3 through its SH2 domain. STAT3 is then phosphorylated at Y705 by non-receptor Janus Kinases (JAKs: JAK1, JAK2, JAK3 and TYK2) or Src kinase, among other possible kinases. This key tyrosine is very conserved among all STATs, relevant to their transcriptional activity, and by far the most studied PTM.
Extensive evidence indicates that STAT3 exists mainly as an unphosphorylated dimer in the cytosol that changes conformation upon Y705 phosphorylation. Tyrosine-phosphorylated STAT3 dimers accumulate in the nucleus fast, in the range of minutes after cytokine stimulation. Once in the nucleus, STAT3 dimers bind to GAS sequences in the DNA and activate the transcription of target genes, such as BCL2, VEGF and MCL-1 [55]. Canonical transcriptional activity can be enhanced, inhibited or fine-tuned by additional PTMs or by the formation of complexes between STAT3 and other transcriptional regulators, such as CBP/p300, Smads, or retinoic acid receptors. Cytokine-stimulated STAT3 activity is usually transient and the signal can be terminated by various intracellular proteins, such as protein tyrosine-phosphatases or Suppressor of cytokine signaling-3 (SOCS3) or PIAS. STAT3 regulates the expression of several genes involved in positive or negative feedbacks for the STAT3 canonical pathway, including STAT3 itself, IL-6 or SOCS3.
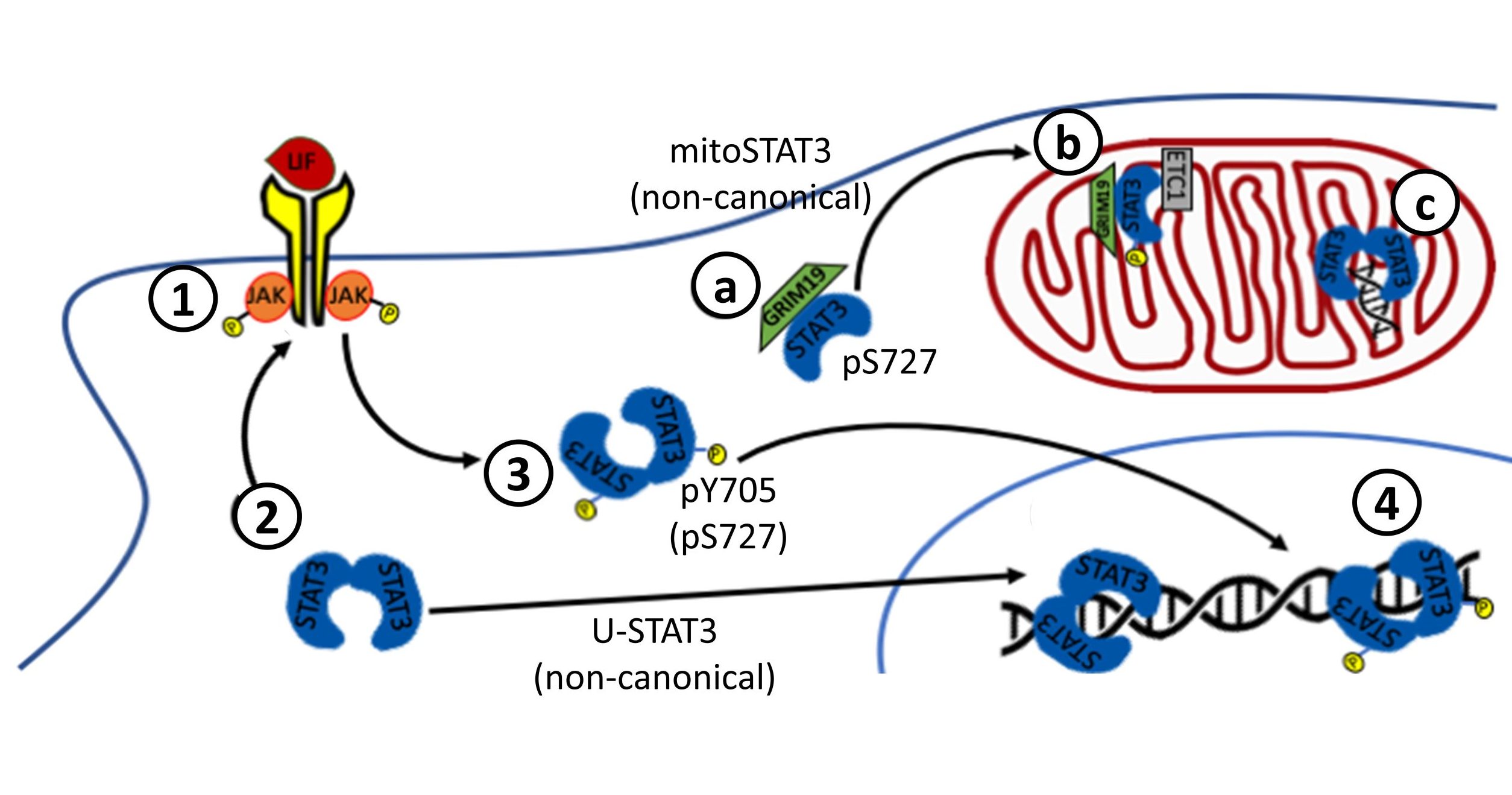
Non-canonical STAT3 pathways
Non-canonical STAT3 pathways have in common that they are not necessarily dependent on Y705 phosphorylation or transcriptional activity of STAT3. Unphosphorylated STAT3 (U-STAT3) can homodimerize, translocate to the nucleus, interact with transcriptional regulators, bind to DNA and regulate the transcription of a distinct set of genes, such as Cyclin B1, E2F1, M-Ras and Met. Nuclear levels of U-STAT3 are directly correlated with the malignancy of glioma, and nuclear accumulation of U-STAT3 is also observed in the development of cardiac hypertrophy.
STAT3 can be also found at the mitochondria, where it can act either as a transcription factor for mitochondrial genes related to the electron transport chain or, most surprisingly, as a direct regulator of the mitochondrial electron transport activity. Although both phosphoY705 and phosphoS727 are detected in mitochondrial STAT3, only S727 phosphorylation is essential for its functions. Mitochondrial STAT3 is involved in tumoral progression.
More recently, STAT3 has been also found in the endoplasmic reticulum, at the mitochondria-associated ER membrane (MAM). STAT3 regulates ER calcium homeostasis by interacting with and facilitating the degradation of the inositol trisphosphate receptor 3, reducing the ER-mitochondria calcium flux and preventing apoptosis. Neither the phosphorylation of STAT3 at Y705 nor S727 is required for both ER localization and IP3R3 interaction but MEFs cells expressing mutated STAT3 at S727 (S727A) a significant increase of calcium release upon ATP stimulation compared with WT and Y705F mutant.
STAT3 is regulated by post-translational modifications
The key role of Y705 phosphorylation in STAT3 canonical functions is undeniable, and the existing evidence strongly indicates that the rest of STATs share this mechanism. STAT3 phosphorylation at Y705 or S727 are most often used as a surrogate of its activation, and new discoveries associated to the STAT3 pathway are frequently presented in relation to these events. However, more than 80 PTMs have been described for STAT3 (Fig. 1D), and there are growing indications of their potential relevance for canonical and non-canonical STAT3 pathways and functions. We are currently exploring 1) which PTMs are constitutive and which are stimulated by cytokines; 2) which PTMs have a redundant function and which are mutually exclusive; and 3) how STAT3 molecules carrying different PTMs or mutations interact with each other. Considering the high number of possible PTMs, we find unlikely that the intracellular population of STAT3 molecules is homogeneously modified, and this probably involves interactions between asymmetrically modified STAT3 molecules. Whether this heterogeneity plays an active role in STAT signaling or is rather a problem the cell must deal with, remains to be elucidated. These and other questions are important gaps in our knowledge of the STAT pathways, which are not as simple and straight-forward as they are commonly depicted. Addressing them could be very relevant to our understanding and manipulation of inflammation, immunity, development, cancer, response to injury or tissue regeneration.
For more information, read our latest articles on the subject:
Diallo M and Herrera F (2021), The role of understudied post-translational modifications for the behavior and function of Signal Transducer and Activator of Transcription 3. Review. FEBS J. https://doi.org/10.1111/febs.16116.
Letra-Vilela R#, Cardoso B#, Silva-Almeida C, Rocha AM, Murtinheira F, Branco-Santos J, Rodriguez C, Martin V, Santa-Marta M, Herrera F (2020). Can asymmetric post-translational modifications regulate the behavior of STAT3 homodimers? FASEB Bioadv. 2020 Jan 27;2(2):116-125. doi: 10.1096/fba.2019-00049.